Cracking the Code within Us: Bioinformatics of the Human Genome
My research group uses computational models to study the functional potential of each of the three billion pairs of chemical bases in the human genome. Ultimately, we are paving the way to designing personalized interventions against disease, which technological advancements are finally pushing toward reality.
The rise of bioinformatics
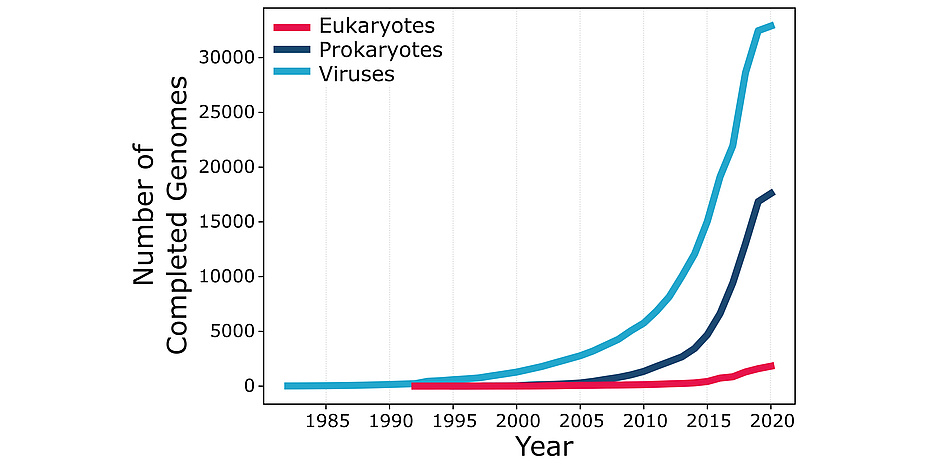
According to the U.S. National Center for Biotechnology Information, bioinformatics “is the field of science in which biology, computer science, and information technology merge into a single discipline”. The origins of bioinformatics can be traced back to the work of Margaret Oakley Dayhoff (1925-1983), a professor at Georgetown University Medical Center who devoted most of her career to the creation and manipulation of biological sequence databases. Bioinformatics received a major thrust from the execution of the Human Genome Project, a publicly funded 13-year-long project initiated in 1990. The HGP determined – sequenced – the three billion pairs of chemical bases in the DNA of the human genome and opened the “big data” era in biology. Further advances in DNA sequencing technologies now make it possible to sequence an entire human genome in only a few days at a tiny fraction of the cost incurred by the HGP. Thousands of genomes of multiple human individuals and many other life forms have been sequenced since the completion of the HGP, with their number growing exponentially and with no end in sight (see Figure 1). This data deluge is changing the face of biology, providing both opportunities and challenges for bioinformatics.
Bioinformatics of gene regulation
Despite the profound impact of the HGP on basic research, we still do not fully know how the genome operates, how it encodes the properties – the phenotype – of an organism.
Nearly all cells in the human body contain the same genome, and thus the same set of genes. Nevertheless, they are able to develop different structures and functions. This is because not all genes are expressed in each cell at a given time. Gene expression can be regulated at any point in the pathway from DNA to RNA to a functional protein. However, for most genes, transcriptional regulation – the conversion of DNA to RNA – is critical. In higher eukaryotes such as humans, transcriptional regulation involves a complex interplay between cis- and trans-regulatory elements and chromatin remodeling. How the precise instructions that determine gene expression patterns are encrypted in the genome remains a central question in biology. This is the focus of my research group (see Figure 2).
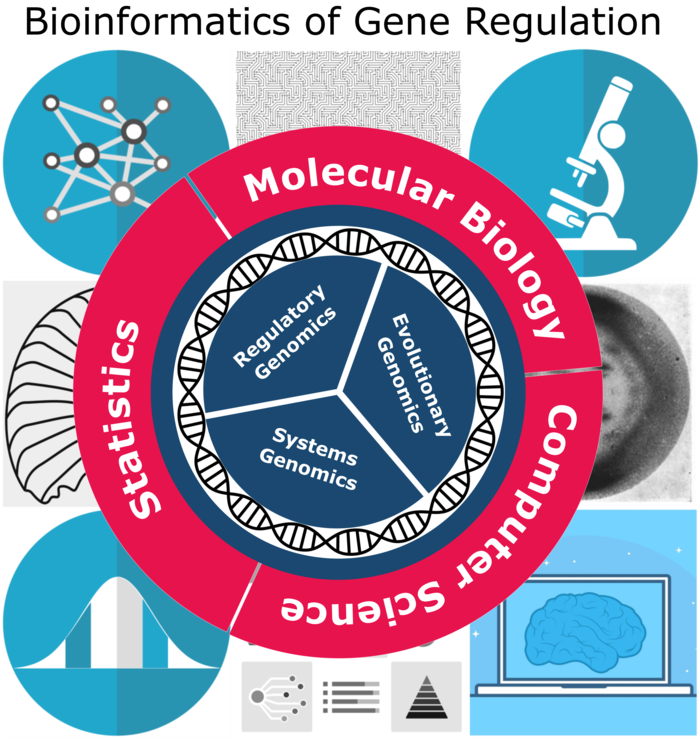
Figure 2: Bioinformatics research at the Institute of Biomedical Informatics.
Using artificial intelligence to annotate the genome
Cis-regulatory elements, such as promoters and enhancers, are short DNA sequences that interact either physically or biochemically with transcription factor proteins to regulate transcription. Transcription requires the assembly of a complex comprising the enzyme RNA polymerase II and many other proteins collectively referred to as “general transcription factors”. The interaction of this complex with the gene promoter is sufficient to initiate transcription at basal levels. However, regulated transcription further requires the interaction of this complex with transcription factors specifically bound to other cis-regulatory elements, which is facilitated by the “mediator” and “cohesion” complexes (see Figure 3).
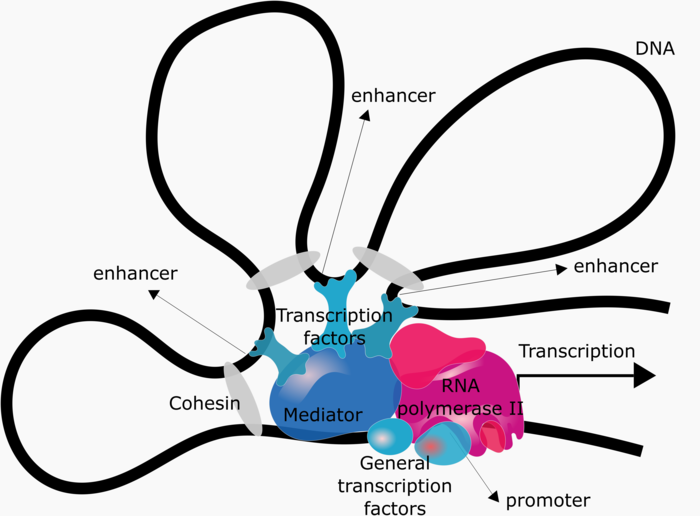
Figure 3: Regulation of gene expression. DNA looping allowing cis-regulatory elements to physically interact and activate gene expression.
The holy grail of regulatory genomics is to uncover a “cis-regulatory code”, which should enable us to predict gene expression based on the sequence of cis-regulatory elements. Cis-regulatory sequences are scattered across virtually the entire genome, making their identification challenging. Furthermore, many cis-regulatory elements are only active under particular conditions. Experimentally testing each stretch of DNA for cis-regulatory activity would require an infinite number of time- and resource-intensive assays. To tackle the problem, we have been using machine learning, a fundamental concept of Artificial Intelligence, for many years.
"Nothing in biology makes sense except in the light of evolution"
In 1973, the geneticist Theodosius Dobzhansky published an essay whose title rapidly became a popular catchphrase. Evolution of organismal properties – the phenotype – results primarily from genomic alterations. Hence, research in the emerging field of evolutionary genomics can provide new insights into human biology and medicine. For example, advancing our knowledge of human evolutionary history would shed light on how populations differ in their genetic risks for common and rare diseases. A theory proposed in the 1970s that is now mainstream proposes that evolution of gene regulation, rather than of the genes themselves, is largely responsible for phenotypic evolution. My research group uses genomic approaches to address long-standing evolutionary questions and evolutionary theory to understand genome structure and function. Specifically, we are investigating how cis-regulatory elements evolve their regulatory activity and how genetic mutations may lead to the disruption of the interactions between cis-regulatory elements and their target genes.
Dysregulation of regulatory networks in disease
During the past two decades, whole-genome sequencing studies have uncovered thousands of variants in the human genome associated with hundreds of diseases. Unfortunately, this knowledge has hardly been reflected in new treatments. First, most of such variants are not located within genes, which makes their interpretation difficult. Second, the susceptibility and pathology of most complex genetic diseases, such as diabetes and multiple sclerosis, are likely to be determined by multiple interacting genetic factors. With the aim of gaining a mechanistic understanding of genetic disease and identifying molecular intervention targets, my research group develops systems genomics approaches integrating sequencing data with biological or clinical information. In contrast to traditional strategies, we infer regulatory networks in which the nodes represent not only genes, but also cis-regulatory elements and more abstract entities, such as molecular functions, biological processes, and pathways, which permit a comprehensive characterization of the system. These networks permit the interrogation of genetic variants of unknown significance that would otherwise have gone unnoticed.
We are now firmly in the big data era of biology. While computational power is currently growing exponentially, the amount of biological data generated is growing even faster. Genome sequencing will soon become an integral part of standard medical diagnosis and treatment. This data deluge will offer tantalizing possibilities for bioinformatics in the field of personalized medicine (see Figure 4).
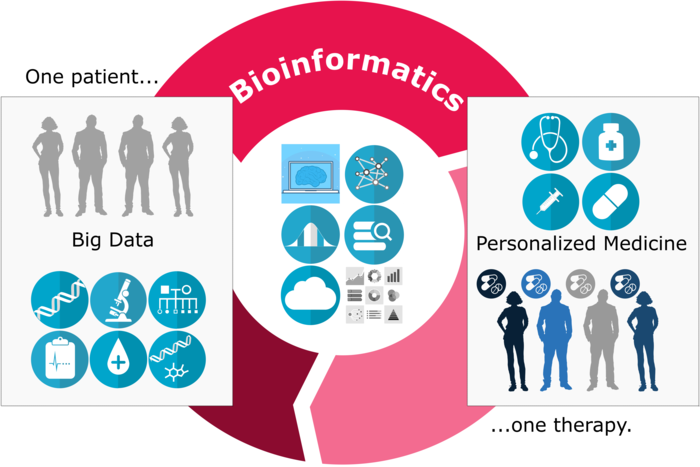
Figure 4: Bioinformatics for personalized medicine: the synergistic cycle of hypothesis-driven and data-driven experimentation.
Kontakt
Leila TAHER
Univ.-Prof. Dr.rer.nat.habil.
Institute of Biomedical Informatics
Stremayrgasse 16/I
8010 Graz
Phone: +43 316 873 5380
leila.taher@tugraz.at